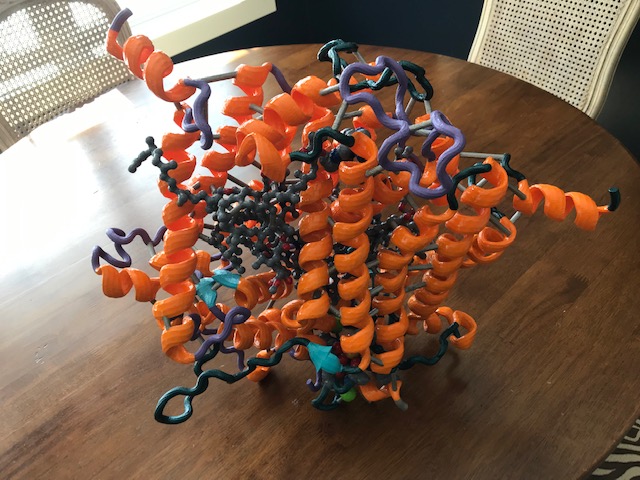
In support of this technology and to further promote what I
feel is a potentially valuable educational tool, I have been building a number of these models and posting them to Thingiverse. Anyone can go to Thingiverse and download the files for printing on a MakerBot or comparable model printer. I have already constructed a model of the FeMo-cofactor, which is the complex metallocenter found in the enzyme nitrogenase, an enzyme I have studied for many years. I’ve also printed two models of the simple proteins Cytochrome C and Plastocyanin. I’ve also broken out the model paints and given them some color. You can find my work under MoleculeMaker on Thingiverse.
To construct our models, the coordinate files are downloaded from the Protein Data Bank as PDB files. We usually clean these up and remove any water molecules. These are then transferred to the program VMD, which is capable of producing an STL file, needed for import by the Makerware program. I usually select something specific from the protein to highlight. For example, I represent the heme molecule in cytochrome C as a CPK model, and then show the protein backbone. At this point, things get complicated, as VMD likes to layer the different representations on top of one another, which causes problems when trying to print. We use the online software Tinkercad to import the files, and then render a smaller file that removes all of the internal scaffolds, resulting in a much nicer print. Tinkercad was recently purchased by Autodesk. I have also tried using SketchUp, but this software has a much steeper learning curve. We are also interested in hearing from others that are using this 3 dimensional printing technology in similar applications, as there may be additional software tools that we are missing.
Several of my protein models have been constructed as they have relevance to the projects we work on in the laboratory. This model of the wax ester synthase enzyme is based on a theoretical structure we utilize in the laboratory for this wax ester synthase enzyme. Since there is currently no published crystal structure for this enzyme, we have to use structures from related enzymes as a “best guess” for what the structure of our enzyme might look like. Using this model, we have predicted where specific residues might reside in our model, and have selected these for site specific mutagenesis studies. These experiments have produced several interesting results which we recently published in the journal Applied and Environmental Microbiology (2013). We also published a paper in 2012 that characterized the differences between five homologs of this enzyme in the same journal. This enzyme is a key focus of study in our laboratory, as it produces an ester product from a fatty acid and a fatty alcohol. If the alcohol is small enough, then the product would be similar to biodiesel. When the enzyme produces the product of the fatty acid and an alcohol such as ethanol, then the product has been termed “microdiesel” by another research group, as it is derived from a microorganism. This enzyme may someday be utilized to produce biodiesel from simple feedstocks such as sugars, or from complex biomass such as cellulose. Most ideally, we would like to produce it in a functioning phototroph, such as a cyanobacterium. The use of the models is thus another tool to be used in our research efforts.
One drawback to our previous 3D prints is the excessive effort that is required to remove the scaffolding to support the molecule during the printing process. Based on some of the original support systems provided as defaults for software like Makerware, the process of removing the supports could take several hours and results in very sore hands and potential damage to the model. Thus, I have been working recently to improve certain models by incorporating our own structural supports. This results in less plastic utilized to print the model, and also drops down the time to remove the structure from several hours to a few minutes. It does require the use of a modeling knife to trim off the supports, but results in a much nicer looking final product.
To the left is the 3D model for the acyl carrier protein as is looks following printing, while the image to the right shows the model 30 minutes later following removal of the supports and some cleanup with a modeling knife to soften the edges. This model is very strong and retains shape better than some of my previous models. I am currently constructing a model of the enzyme quercetinase as well as a model of the substrate. This is a nice enzyme to do modeling with, as the substrate is large and the structure was solved with a substrate bound. My hope is to build a model that students can use to insert the substrate component into the active site, which will assist in their understanding of how enzymes orient substrates to improve catalysis. We also utilize the quercetinase enzyme as part of a teach laboratory experiment in our BBE3013 course at UMN.